Raw, purified DNA can be used to analyze sequence data, microsatellites, and more. My phylogeography project uses sequence data from a mitochondrial gene and several nuclear introns. These stretches of DNA sequence range from 500 to 1000 base pairs in length. In order to sequence them, the raw DNA must be chopped into the desired fragments (loci) and amplified by many orders of magnitude. The process that accomplishes this is called PCR: Polymerase Chain Reaction. Prepare your brains for the biochemical heavy lifting of genetic work.
The raw DNA is added to a mixture of the following:
dNTP (deoxyribonucleotide triphosphate) - the nucleotides used to compose the new strands
primers - short sequences of DNA that bind to either end of the locus to be copied
polymerase - the enzyme that binds to the primers and creates the new strands of DNA
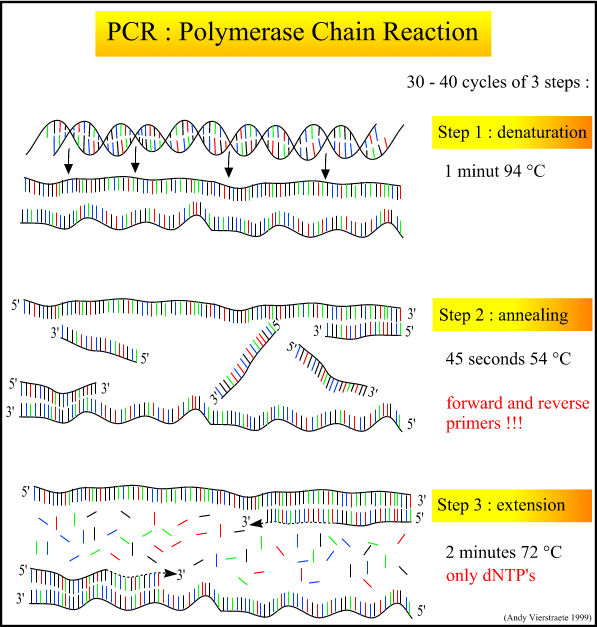
The PCR process itself is a series of cycles, each cycle composed of three steps at a different temperature. The first step heats the mixture to roughly 95 degrees C, denaturing and separating the parent DNA strands. The second step cools the mixture to roughly 54 degrees C, and allows the primers and polymerase to bind. A special polymerase, Taq, is needed to withstand the high heat of denaturation without denaturing itself. Taq comes from the heat-tolerant bacteria Thermophilus aquaticus, discovered in Yellowstone hot springs in the 1960's. The third step is intermediate in temperature (72 degrees C), allowing the polymerase to create the new strands by adding nucleotides off the end of the primer.
dNTP (deoxyribonucleotide triphosphate) - the nucleotides used to compose the new strands
primers - short sequences of DNA that bind to either end of the locus to be copied
polymerase - the enzyme that binds to the primers and creates the new strands of DNA
The PCR process itself is a series of cycles, each cycle composed of three steps at a different temperature. The first step heats the mixture to roughly 95 degrees C, denaturing and separating the parent DNA strands. The second step cools the mixture to roughly 54 degrees C, and allows the primers and polymerase to bind. A special polymerase, Taq, is needed to withstand the high heat of denaturation without denaturing itself. Taq comes from the heat-tolerant bacteria Thermophilus aquaticus, discovered in Yellowstone hot springs in the 1960's. The third step is intermediate in temperature (72 degrees C), allowing the polymerase to create the new strands by adding nucleotides off the end of the primer.
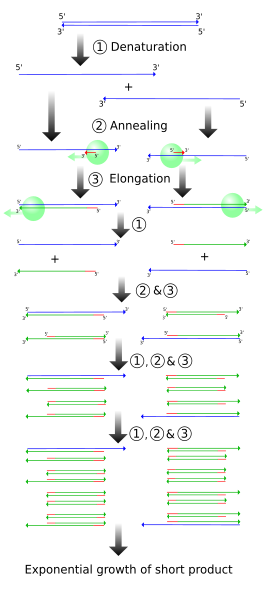
The simplified figure above demonstrates PCR starting with a set-length strand. However, we do PCR on raw DNA. When raw DNA is subjected to PCR, the entire genome is added and only ~1000 base pairs (bp) need to be copied.
How does PCR achieve this? The following figures demonstrates (cropped from the wikipedia source).
One single cycle of the above steps generates two open-ended DNA strands. Each contain the parent strand, along with a new strand that starts with the primer and runs until the step ends. This new strand can be thousands of bp long, depending on how long the extension step is run. Note that the two newly formed strands of DNA are complementary.

Another cycle is completed, generating more open-ended copies from the parent strand. On the open-ended primer strands from cycle 1, the opposite primer binds and adds nucleotides until it reaches the first primer, resulting in a correct-length strand bound to an open-ended strand.

The third cycle creates more open-ended/parent and open-ended/correct-length strands, along with the first replication of the correct-length double strands. Each further cycle allows the number of gene copies to grow exponentially.
How does PCR achieve this? The following figures demonstrates (cropped from the wikipedia source).
One single cycle of the above steps generates two open-ended DNA strands. Each contain the parent strand, along with a new strand that starts with the primer and runs until the step ends. This new strand can be thousands of bp long, depending on how long the extension step is run. Note that the two newly formed strands of DNA are complementary.

Another cycle is completed, generating more open-ended copies from the parent strand. On the open-ended primer strands from cycle 1, the opposite primer binds and adds nucleotides until it reaches the first primer, resulting in a correct-length strand bound to an open-ended strand.

The third cycle creates more open-ended/parent and open-ended/correct-length strands, along with the first replication of the correct-length double strands. Each further cycle allows the number of gene copies to grow exponentially.

Putting it all together, we get the following. Sorry about the poor quality of the cropping, but it was the best representation I could find.
These temperature cycles are generated by a Thermocycler. Up to 96 samples are loaded, the exact temperature/time cycle set, and the Thermocycler automates the whole process with tight temperature control. I generally run my samples for about 30 cycles, generating about 2^30 copies of the loci I am sequencing in about 3 hours.



When done, I just run a check with gel electrophoresis to test whether the PCR was successful. I label a small sample of the DNA with dye, run it out on an agarose gel using electrical current (DNA has a negative charge), and view it with UV to highlight the dye. If the PCR was successful, a single bright band will show up of the appropriate length.
UV imaging

If all goes well, I end up with this, rows and rows of PCR product:

With PCR product in hand, literally billions of copies of a locus, we can now sequence it.
Part 4: From DNA to Data (2)
No comments:
Post a Comment